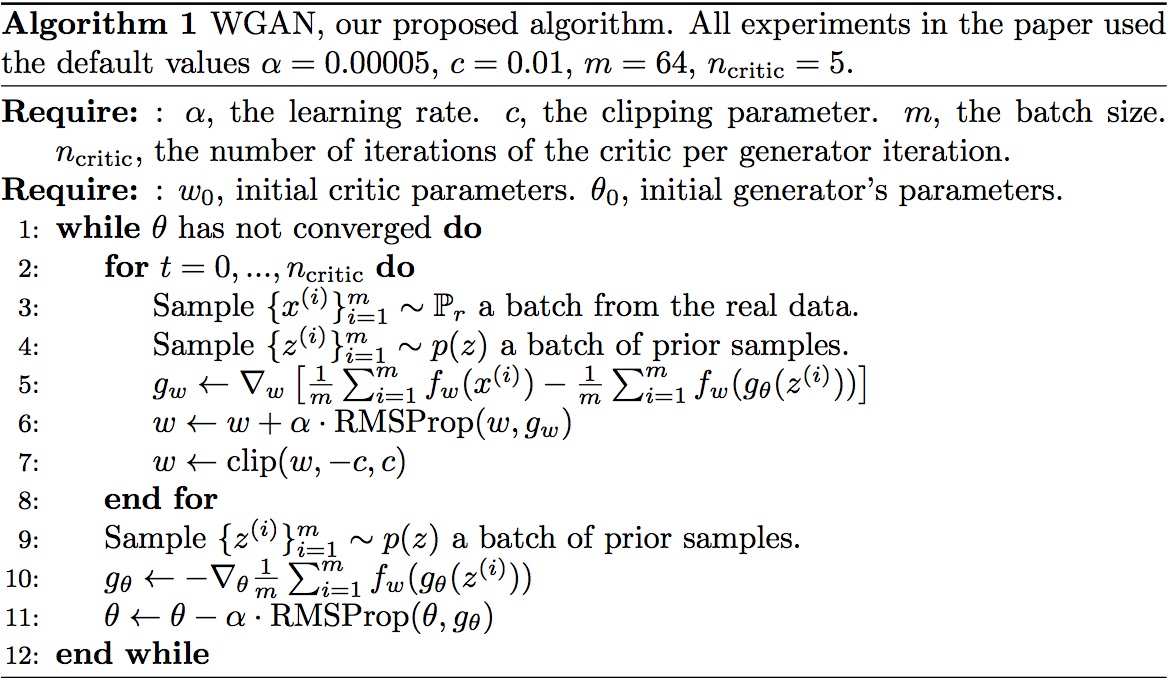
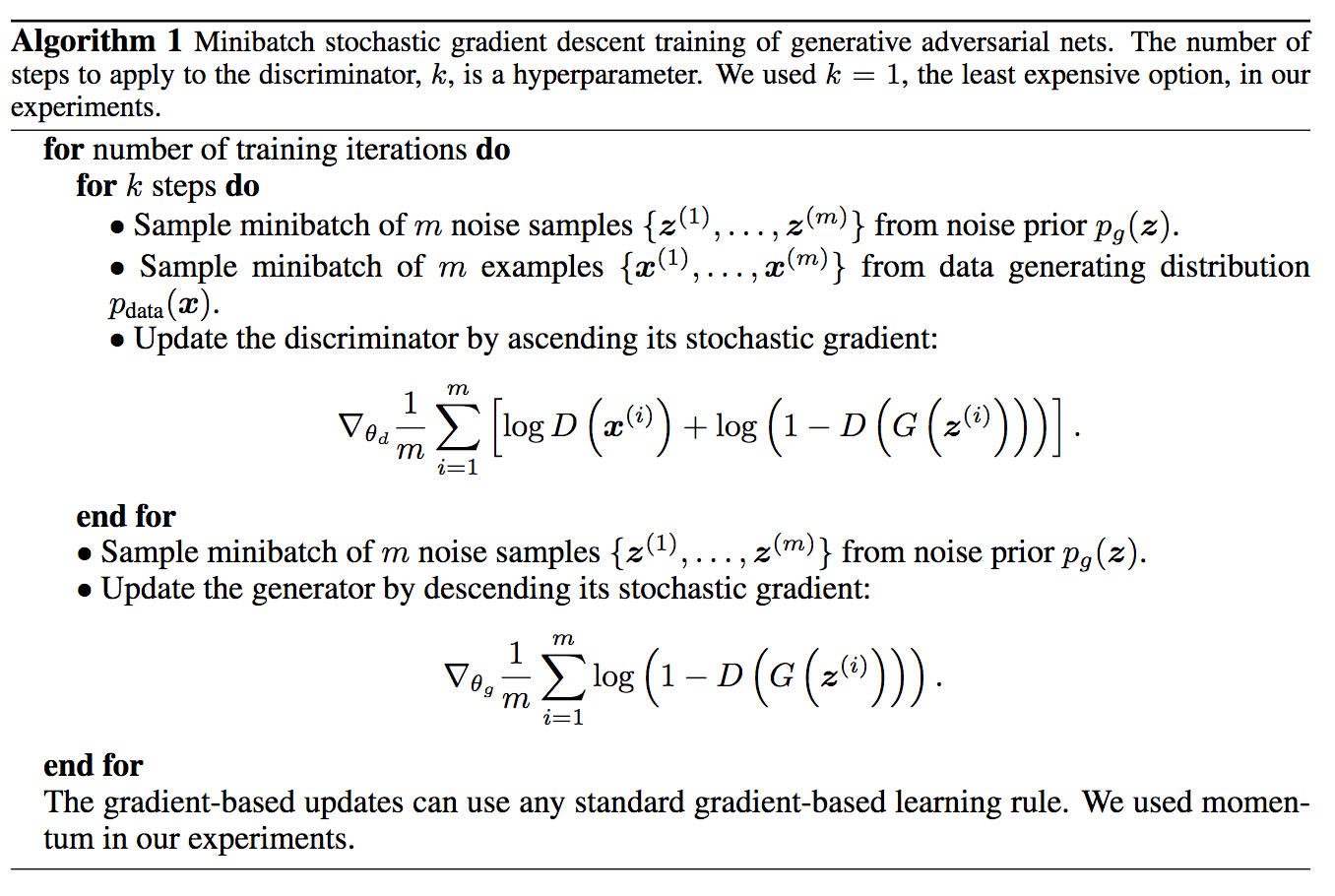
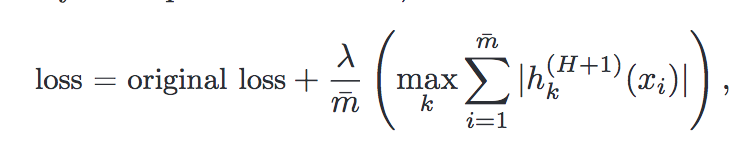In 2019, Google published a new Python library called “tensornetwork” (arXiv:1905.01330) that facilitates the computation of… tensor networks. Tensor network is a tool from quantum many-body theory, widely used in condensed matter physics. There have been a lot of numerical packages for tensor computation, but this library takes it to the next level because of its distinctive framework.
What is a tensor network, though?
“A tensor network is a collection of tensors with indices connected according to a network pattern. It can be used to efficiently represent a many-body wave-function in an otherwise exponentially large Hilbert space.”
https://www.perimeterinstitute.ca/research/research-initiatives/tensor-networks-initiative
Renormalization Group (RG)
It is not until recently that tensor networks have its application in machine learning. As stated in a previous post, a mathematical connection between restricted Boltzmann machine (RBM) and variational renormalization group (RG) was drawn. (arXiv:1410.1831) It shedded light to the understanding of interpretability of deep learning, which has been criticized to be a black box. However, RBM is just a type of unsupervised machine learning, but how about others?
Seeing this, Schwab, one of the authors of the RG paper, and Stoudenmire did some work to realize the use of RG in machine learning. Stoudenmore is a physicist, and he made use of density matrix renormalization group (DMRG) that he is familiar with, and invented a supervised learning algorithm, which is later renamed tensor network machine learning (TNML). The training is adapted from the sweeping algorithm, the standard of DMRG, that combining bipartite site one-by-one, updating it, and decomposing into two site by studying its quantum entanglment (using singular value decomposition, or Schmidt decomposition).

Instead of bringing interpretability to deep learning, this work in fact opened a new path of new machine learning algorithms with known techniques.
What is RG?
Renormalization group (RG) is a formalism of “zooming out” in scale-invariant system, determining which terms to truncate in a model. It is an important formalism in high energy physics and statistical field theory. (See Ma’s book for reference.)
Density matrix renormalization group (RG) is a variational real-space numerical technique that look at collections of quantum bits (zoomed-out) as a block. It was invented by Steven White, and it is useful in studying strongly correlated electronic systems. (PRL 69 (19): 2863-2866 (1992)). However, the original DMRG paper is not very accessible, until it is rephrased using the tensor network notation (TNN), as shown in Schollwoeck’s article.
Is Tensor Network Related to Quantum Computing?
This is not an easy question to answer. Tensor networks come from quantum physics, but quantum physics is usually not directly leading to quantum computing. In fact, classical computing hardwares have a lot of quantum physics in it. A simple answer to this question is no, as the algorithm using tensor network is implemented in classical computers.
There have been a lot of publications on quantum machine learning lately. A classic book on this topic is written by Peter Wittek. The book covers topics on basic machine learning and quantum computing, and then quantum machine learning algorithms. There is a quantum counterpart of each of the common machine learning algorithms in the book. However, we know it would be much more useful if there are new algorithms exploiting the advantages of quantum computing. Tensor network is a natural choice as it builds on qubits, and the representations and operations are naturally quantum.
Next…
Tensor network is an interesting subject from both a theoretical and applicational perspective. In coming posts I will talk about its application on machine learning and a taste of codes.
- Github: google/TensorNetwork. [Github] [RTFD]
- Chase Roberts, Ashley Milsted, Martin Ganahl, Adam Zalcman, Bruce Fontaine, Yijian Zou, Jack Hidary, Guifre Vidal, Stefan Leichenauer, “TensorNetwork: A Library for Physics and Machine Learning,” arXiv:1905.01330 (2019). [arXiv]
- “Google TensorNetwork Library Dramatically Accelerates ML & Physics Tasks,” Syncedreview. (2019) [Medium]
- Chase Roberts, “Introducing TensorNetwork, an Open Source Library for Efficient Tensor Calculations,” Google AI Blog. (2019) [GoogleAIBlog]
- “Tensor Networks and Density Matrix Renormalization Group,” Everything About Data Analytics. (2016) [WordPress]
- P. Mehta, D. J. Schwab, “An exact mapping between the Variational Renormalization Group and Deep Learning,” arXiv:1410.3831 (2014). [arXiv]
- Sheng-kang Ma, Modern Theory of Critical Phenomena, (New York, NY: Routledge, 2018). [Amazon]
- S. R. White, “Density matrix formulation for quantum renormalization groups,” Phys. Rev. Lett. 69, 2863 (1992). [APS]
- Ulrich Schollwoeck, “The density-matrix renormalization group,” Rev. Mod. Phys. 77, 259 (2005); arXiv:cond-mat/0409292. [arXiv]
- Ulrich Schollwoeck, “The density-matrix renormalization group in the age of matrix product states,” Annals of Physics 326, 96 (2011); arXiv:1008.3477. [arXiv]
- Peter Wittek, Quantum Machine Learning: What Quantum Computing Means to Data Mining (San Diego, CA: Academic Press, 2014). [Amazon] [PDF
]
- Jacob Biamonte, Peter Wittek, Nicola Pancotti, Patrick Rebentrost, Nathan Wiebe, Seth Lloyd, “Quantum Machine Learning,” Nature 549, 195-202 (2017). [Nature]
- Tensor Networks: From Entangled Quantum Matter to Emergent Space Time, Perimeter Institute. [Perimeter]
Feature picture taken from Perimeter Institute.